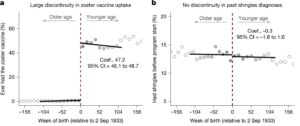
When we talk about music, we are almost automatically talking about variation: diverse kinds of music and different levels of musical ability come to mind. But what about love of music? Sure, some people enjoy music more than others, but I didn’t know that this variation is vast and that it seems to have heritable components until I read a new paper in Nature Communications. The authors used twin studies and cool objective measures of music enjoyment to explore the genetic influences on how people come to love (enjoy) music to varying extents. The story is complicated, thrillingly or perhaps frustratingly so, but the fact is that about half of the variation in musical enjoyment results from genetic effects. I thought my kids like good folk rock because of our family environment, but it seems that DNA was involved too.
I like folk rock but I also really like great scientific writing, and this paper has some chef’s kiss moments. In their Introduction, they call out the great Oliver Sacks, then deftly refer back to his description a bit later. Here:
This human attraction to music has always been considered somewhat baffling and mysterious, leading many to ask why music has such power over humans. Oliver Sacks highlighted this conundrum in the opening of his beautifully written commentary, The Power of Music: “What an odd thing it is”, he wrote “ to see an entire species—billions of people—playing with listening to meaningless tonal patterns, occupied and preoccupied for much of their time by what they call ‘music’”.
Two paragraphs later they write:
A better understanding of such genetic effects will elucidate how the ability to enjoy music is passed from one generation to the other, clarify the mechanisms linking genotypes, brains, and affect, and contribute to solving the conundrum of how “meaningless tonal patterns” can have such a powerful effect on humans.
Clear, effective writing is almost as good as music. Almost.
Twin modelling reveals partly distinct genetic pathways to music enjoyment
In Nature Communications, 25 March 2025
From the groups of Fredrik Ullén, Simon E. Fisher, and Miriam A. Mosing
Snippet by Stephen Matheson